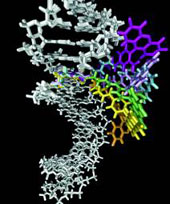
Simulation of biomolecular dynamics and thermodynamics
Our work seeks to understand molecular interactions at the atomic level, through the use of electronic structure theory, empirical potentials and molecular simulation techniques. In addition to hybrid QM/MM models, we also have explored more empirical approaches to modelling biomolecular recognition. For example, we have used a combined molecular mechanics and continuum solvent approach to calculation of thermodynamics, leading to new insights into the recognition of oligosaccharides by the protein, concanavalin A.
We have used a similar approach to predict fluorophore-DNA interactions pertinent to molecular diagnostics. This method has been extended to evaluate the free energies of binding of closely-related ligands, with a view to application in rational drug design.
Selected publications
- Ab initio protein folding using a cooperative swarm of molecular dynamics trajectories. N. J. Bruce and R. A. Bryce. J. Chem. Theory Comput. 2010, 6, 1925 – 1930.
- Assessment of QM/MM scoring functions for molecular docking to HIV-1 protease. P. Fong, J. P. McNamara, I. H. Hillier and R. A. Bryce J. Chem. Inf. Model. 2009, 49, 913-924.